Public Channels
- # anonymous-question-box
- # ask-the-admin
- # day01-intro-genomics-biometric-model
- # day02-sem-study-design-phenomics
- # day03-quality-control-gwas
- # day04-heritability-and-gcta
- # day05-meta-analysis-prs
- # day06-genomicsem
- # day07-causal-modeling-mendelian-randomization
- # day08-sequencing-introduction-to-hail
- # day09-rare-saige
- # day10-pathway-and-gene-based-analyses
- # documentation
- # f-abcd-repronim-course-videos-instructions
- # f-creation-of-videos
- # general
- # python-intro
- # random
- # session-a
- # session-b
- # technical-support
Private Channels
Direct Messages
Group Direct Messages
@Jeff Lessem (he/him) has joined the channel
@Michael Neale has joined the channel
@Elizabeth Prom-Wormley has joined the channel
@Rob Kirkpatrick has joined the channel
@Sophie van der Sluis has joined the channel
@Test Student has joined the channel
@Sarah Brislin (she/her) has joined the channel
@Katie Bountress has joined the channel
@Peter Tanksley has joined the channel
@Charlotte Viktorsson has joined the channel
@Matthieu de Hemptinne has joined the channel
@Sam Freis (she/her) has joined the channel
@Stephanie Zellers (she/her/hers) has joined the channel
@Zoe Schmilovich has joined the channel
@Olivia Rennie has joined the channel
@Christina Sheerin has joined the channel
@William McAuliffe has joined the channel
@Francis Vergunst (he/him) has joined the channel
Any advice for setting up our local R environment to be able to run the scripts from today? I am having some version conflicts that are keeping me from using the preferred “NPSOL” optimizer.
*Thread Reply:* Instructions for installing OpenMx with NPSOL are at https://openmx.ssri.psu.edu/installing-openmx
*Thread Reply:* Thanks, Jeff!
I am still running into some issues, even after installing the NPSOL compiled version. Any thoughts? (R v4.1.0) ```> source('https://vipbg.vcu.edu/vipbg/OpenMx2/software/getOpenMx.R') You are now installing the latest version of OpenMx, compiled with NPSOL, that is available for R-4 Warning in install.packages : unable to access index for repository https://openmx.ssri.psu.edu/software/bin/macosx/contrib/4.1: cannot open URL 'https://openmx.ssri.psu.edu/software/bin/macosx/contrib/4.1/PACKAGES' Warning in install.packages : package 'OpenMx' is not available as a binary package for this version of R
A version of this package for your version of R might be available elsewhere, see the ideas at https://cran.r-project.org/doc/manuals/r-patched/R-admin.html#Installing-packages
> require(umx) Loading required package: umx Loading required package: OpenMx OpenMx may run faster if it is compiled to take advantage of multiple cores. For an overview type '?umx'
Attaching package: 'umx'
The following object is masked from 'package:stats':
loadings
> umxsetoptimizer("NPSOL") Error in umxsetoptimizer("NPSOL") : The Optimizer 'NPSOL' is not legal. Legal values (from mxAvailableOptimizers() ) are:'CSOLNP' and 'SLSQP'```
*Thread Reply:* Just reading through the error message again. Looks like I need a different version of R?
*Thread Reply:* The easiest thing to get through the practical is to just change the optimizer with something like
umx_set_optimizer("SLSQP")
You can also try the install from in umx, so first make sure umx is loaded
library(umx)
install.OpenMx("NPSOL")
It is also possible there is not yet a R 4.1.0 version of NPSOL for Mac yet.
*Thread Reply:* I’ve had similar problems with installing NPSOL before, but I can’t remember how I got around them (I’m on a mac btw). As Jeff noted, easiest thing is to use a different optimizer. They give virtually the same answers except in specific situations (e.g., NPSOL is required for fitting models with nonlinear constraints, which is why I had to install it). Long & short = if you don’t NEED NPSOL, don’t use it. It was needed today bc Rob was going to do something requiring it in his 3rd prac
*Thread Reply:* My practical3 script runs now without NPSOL. It's /faculty/hmaes/2021/day2/practical3/mulACE.R . Also, you do NOT need NPSOL to fit a model with nonlinear constraints. SLSQP is adequate for that purpose almost all the time, and CSOLNP is adequate some of the time.
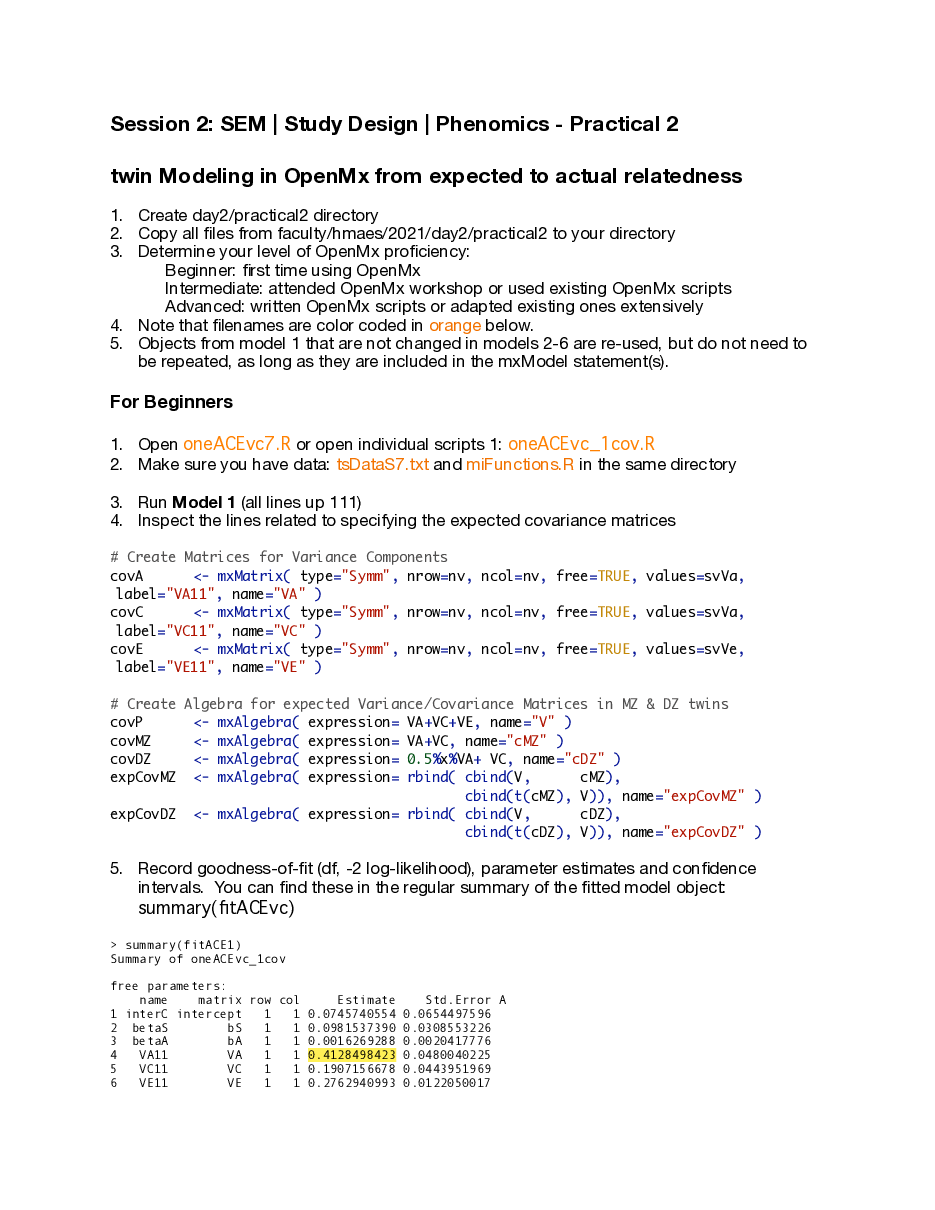
A couple of comments on session 2's practical 2:
The beginner scripts (oneACEvc7.R or oneACEvc7_withQuestions.R) run fine.
The intermediate scripts (oneACEvc72fill.R or oneACEvc72fill_withQuestions.R) may crash Rstudio IF you have not fixed the missing piece AND IF you're using the NPSOL optimizer. They run fine if you do either of the following steps
- You fix the error in the script: correction of line 79: covDZ <- mxAlgebra( expression= 0.5%x%VA+ VC, name="cDZ" ) This error in the script should give you wrong results but shouldn't crash Rstudio. We'll look into fixing that bug!
- If you use the SLSQP optimizer by uncommenting line 22:
mxOption(NULL,"Default optimizer","SLSQP")
(and commenting line 20). I have updated the miFunctions.R script so it doesn't overwrite the optimizer you chose in your script.
If you have any further questions on the session, don't hesitate to post them in slack or email us (hmaes@vcu.edu). I'm also happy to setup an additional zoom session with anyone interested to go over the practical in more detail!
I think I know what the problem was with NPSOL during Session A. R, on IBG's RStudio Server, was apparently compiled with OpenBLAS. There is a known issue with NPSOL and tuned BLAS/LAPACK implementations: https://openmx.ssri.psu.edu/comment/8445#comment-8445
@Jeff Lessem (he/him) has renamed the channel from "sem-study-design-phenomics" to "day01-sem-study-design-phenomics"
@Jeff Lessem (he/him) has renamed the channel from "day01-sem-study-design-phenomics" to "day02-sem-study-design-phenomics"
I'm in putty, trying to copy and paste today's practical files from tutor's directory to my home directory. I'm getting the following problem: workshop-40:~/Day2> cp /faculty/hmaes/2021/day2/practical1** . cp: -r not specified; omitting directory '/faculty/hmaes/2021/day2/practical1'
*Thread Reply:* It's complaining you left the -r option off cp . The -r option tells it to copy a whole directory, and it's complaining it won't do that without the -r.
*Thread Reply:* Also, make sure you get the space between the two arguments:
cp -r /faculty/hmaes/2021/day2/** ./
*Thread Reply:* I have the files now - thank you.
add -r in front. I'm so sorry, I thought I had corrected this.
cp -r /faculty/hmaes/2021/day2/** ./
It would be a bit safer to cp -r -i /faculty/hmaes/2021/day2/** ./ this will ask whether to overwrite in case the same files already exist in the copying-to current directory (./)
Group 5: terribly sorry to leave you so abruptly 45 minutes ago! Zoom locked up on me.
> library(umx) # load the umx library > selDVs=c("phenoT") # the name of the phenotype phenoT1, phenoT2 > ADEmodel_1=umxACEv(
- name='ADE',
- selDVs=selDVs, # the phenotype
- selCovs=NULL, # fixed covariates ... none here
- dzData=dataDZ, # the DZ data frame
- mzData=dataMZ, # the MZ data frame
- sep='', # phenoT""1 = phenoT1, phenoT""2 = phenoT2
- dzCr=.25 ) # in the ACE model dzCr=1, in the ADE model dzCr=.25 Running ADE with 4 parameters Error incurred trying to run model: model = mxTryHard(model) might help? NPSOL is not available in this build. See ?omxGetNPSOL() to download this optimizer
*Thread Reply:* Was this running on the workshop R server or on your local machine?
while running the above code from Conor's workbook today, i got the error message at the bottom. any suggestions?
Hi Kirk: that is weird! " NPSOL not available in the build " means that you installed OpenMx with R (rather than installing from the OpenMx site). Perhaps you included mxOption(NULL, "Default optimizer","NPSOL")
mxOption(NULL, "Default optimizer","CSOLNP")
Was this running on the workshop R server or on your local machine?
Perhaps your code includes the line mxOption(NULL, "Default optimizer","NPSOL") If you replace this with mxOption(NULL, "Default optimizer","CSOLNP") the problem might go way.
Thanks Conor, I'll try that. And you are right - I installed OpenMx with R, and Sarah, I was running on my local PC.
ONLY FOR YOUR OWN COMPUTERS (ie don't do this on the workshop R-studio server)
If you want to install the version of OpenMx which uses the NPSOL optimizer,
copy the following line into the R command line and press return.**source**('<https://vipbg.vcu.edu/vipbg/OpenMx2/software/getOpenMx.R>')
The demo script for practical 3 from day 2 now runs on the RStudio Server. It's /faculty/hmaes/2021/day2/practical3/mulACE.R
I was going through the examples once more, and I tried to extend them to include covariates.
Questions:
1: is residualizing (i.e., umx_residualize()) first the phenotype on other variables (i.e., age, sex), and fitting a biometric model on the phenotype to estimate variance components equivalent to introducing covariates in the model? If so, how would you test the significance of the covariates (usually, you can try it by removing them from the model and check the model fit, right?)
2: Is that the case for multivariate models too (i.e., residualize J phenotypes on the same Z variables and then fit a multivariate model...)
3: If the covariate is not significant (i.e., age), should I remove it? Or alternatively, assuming I want to “control” for its effect, should I keep the covariate in regardless. What would I do in a multivariate scenario where age is a significant covariate for a variable i and not a variable k?
Additional question: I saw some papers including interaction (age**sex) and quadratic terms (age^2) as additional covariates. Should I always think to test them in my models as good practice?
thanks in advance and apologies for the amount of questions 😅
*Thread Reply:* heads up about this question to @Michael Neale @Hermine Maes @Rob Kirkpatrick
*Thread Reply:* 1. It is equivalent for continuous, multivariate normal data. It is NOT equivalent for ordinal or binary measures. In these latter cases, including covariates in the model is correct, but regressing out ahead of time is not.
- Yes, response to 1 applies in multivariate case
- It is possible to fix to zero those betas that don’t achieve significance. Usually in a twin study the sample size will be large (500+ people). If a regression with N=500 is not significant, its effect size is likely very small so it will not affect results much if it is included or not.
- It might be worth it to include quadratic or interaction effects, but these are unlikely to be significant if the main effects are not. That is because precise counterbalancing of effects is needed to yield sig interactions in the absence of main effects, which asymptotically becomes infinitely unlikely. Visual inspection of the data may suggest quadratic effects, so this can be a useful guide as well.
*Thread Reply:* thank you very much!
*Thread Reply:* One correction to Mike's reply to #1--it is NOT equivalent for continuous multivariate data, either, though I doubt it makes much difference most of the time in practice. I explain why here: https://openmx.ssri.psu.edu/comment/6485#comment-6485
*Thread Reply:* (To clarify: it's not exactly equivalent, but only approximately equivalent.)
*Thread Reply:* Here is my advice: 1) try to avoid two step procedures (first residualizing and then proceeding with the analysis of interest). If it bad practice because the info you use in step one (e.g., phenotype regresses on age) takes the form of parameters (regression coefficient), which come with standard errors (treating parameter estimates as true value is not nice); 2) if you fit a (say) twin model and you are interested in the ACE or ADE components of you phenotype, corrected for a given covariate (age / sex), do not statistically test the covariate (that test is not of interest). Suppose you set alpha=0.05, and you test the significance of the covariate age, and you find a p value of .06.... you decide not te test. The reviewer of your paper may ask: what about the power to detect the age effect (a can of worms).
*Thread Reply:* Thanks. Advice 2) is pretty useful, especially when the effect of the covariate per se is not of interest. 👍
Hello! The linear regression and GCSM lecture for this day says that the R code for lecture examples is in the slide notes, but I don't see slide notes in the posted pdf. Is this posted in a place that I'm not finding? Thanks!
Quick Q for @Hermine Maes, but open also to others. How do we cite/acknowledge in research the code shared? I am explicitly referencing code from this day and to https://hermine-maes.squarespace.com/, but this question potentially extends to when we will use and adapt other scripts shared during the workshop. Thanks in advance 🙂
*Thread Reply:* Hi Giacomo
I don’t know if Hermine has anything more specific, but you could cite the relevant url, and perhaps the output of the citation(“OpenMx”) command in R, which is a bit voluminous but here goes anyway: To cite package ‘OpenMx’ in publications use:
Michael C. Neale, Michael D. Hunter, Joshua N. Pritikin, Mahsa Zahery, Timothy R. Brick Robert M. Kirkpatrick, Ryne Estabrook, Timothy C. Bates, Hermine H. Maes, Steven M. Boker. (2016). OpenMx 2.0: Extended structural equation and statistical modeling. Psychometrika, 81(2), 535-549. doi:10.1007/s11336-014-9435-8
Pritikin, J. N., Hunter, M. D., & Boker, S. M. (2015). Modular open-source software for Item Factor Analysis. Educational and Psychological Measurement, 75(3), 458-474
Hunter, M. D. (2018). State space modeling in an open source, modular, structural equation modeling environment. Structural Equation Modeling, 25(2), 307-324. doi: 10.1080/10705511.2017.1369354
Steven M. Boker [aut], Michael C. Neale [aut], Hermine H. Maes [aut], Michael J. Wilde [ctb], Michael Spiegel [aut], Timothy R. Brick [aut], Ryne Estabrook [aut], Timothy C. Bates [aut], Paras Mehta [ctb], Timo von Oertzen [ctb], Ross J. Gore [aut], Michael D. Hunter [aut], Daniel C. Hackett [ctb], Julian Karch [ctb], Andreas M. Brandmaier [ctb], Joshua N. Pritikin <jpritikin@pobox.com> [aut, cre], Mahsa Zahery [aut], Robert M. Kirkpatrick [aut], Yang Wang [ctb], Ben Goodrich <goodrich.ben@gmail.com> [ctb], Charles Driver <driver@mpib-berlin.mpg.de> [ctb], Massachusetts Institute of Technology [cph], S. G. Johnson [cph], Association for Computing Machinery [cph], Dieter Kraft [cph], Stefan Wilhelm [cph], Sarah Medland [cph], Carl F. Falk [cph], Matt Keller [cph], Manjunath B G [cph], The Regents of the University of California [cph], Lester Ingber [cph], Wong Shao Voon [cph], Juan Palacios [cph], Jiang Yang [cph], Gael Guennebaud [cph] and Jitse Niesen [cph]. (2021) OpenMx 2.19.5-1 User Guide.
BibTeX entries for LaTeX users are obtained by:
toBibtex(citation(‘OpenMx’))
OpenMx was developed with support from NIH/NIDA grants:
R37-DA018673, R25-DA026119, R21-DA024304 To see these entries in BibTeX format, use ‘print(<citation>, bibtex=TRUE)’, ‘toBibtex(.)’, or set ‘options(citation.bibtex.max=999)’.
*Thread Reply:* thank you! 👍
*Thread Reply:* I’ve used the following citation for the website: Maes HH. OpenMx Scripts. 2018; Available from: http://hermine-maes/squarespace.com, but rules may vary by publication.
I’ve been going through the material again with some colleagues, and we hope it’s ok to still ask some questions. Thank you in advance!
1/4 When running a multivariate twin modelling, is there any cutoff (both lower and upper thresholds) for the phenotypic correlation between the study phenotypes?
@Hermine Maes @Charlotte Viktorsson
*Thread Reply:* Hi Ana
No cutoff for lower limit. Usually multivariate studies focus on positively correlated traits, but when more than one variance component is involved, it is possible that one will contribute positively to covariance and the other negatively. The end result is little phenotypic correlation at all. Indeed, this could happen for two traits who share 100% of their causal influences(!) but they still correlate zero.
For upper limits, they don’t exist either, but as the correlation approaches 1, so the variance components of the two traits become identical (and have the same sign effect on the two variables). Some numerical instabilities may occur, especially with model specifications that force ‘sensible’ correlation matrices on the latent factors. Here “sensible” refers to non-negative definite covariance matrices (i.e., ones that don’t have correlations > 1 in absolute value, or inconsistencies (variable 1 and 2 correlate .999 but one correlates with variable 3 .2 and the other .8, say).
HTH!
*Thread Reply:* Thank you, Michael, this is very helpful!
2/ 4 When testing the ADE model, does it make sense to test a DE model, and is it plausible to get D > A estimates?
@Hermine Maes
*Thread Reply:* 2/4 No, the DE model is biologically implausible. See the biometrical genetics lecture from Ben, it is very unlikely that at every trait relevant locus aa and AA genotypes have the same average phenotypic value in the population, while the Aa genotypes differ. Furthermore all these loci need to have p=q=.5 allele frequency, since: VA = 2pq [a + (q -p)d]^2 VD = (2pqd)^2 where p and q are allele frequencies p=1-q, a is the distance from the midpoint of the two homozygotes and d is the heterozygotes’ deviation from the mean. So you can end up with VA>0 unless: i) a=0, ii) p=q, and iii) d > 0. These three together don’t seem to happen often in nature, and for a polygenic trait to involve only such loci seems infinitesimally unlikely.
*Thread Reply:* 3/4 This can and does happen (I think you mean “both A and C are not significant,..“) Essentially, the data collected are sufficient to conclude that there is significant familial resemblance, but that there aren’t enough data points to discern whether that is due to A alone, C alone, or both.
*Thread Reply:* 4/4 Try to figure out WHY the assumptions are violated. Quite often, IME, such model failures are due to failure of the multivariate normality assumption. Possibly, a switch to ordinal data methods will help to avoid the continuous data assumptions.
*Thread Reply:* One other thing about 2/4. WIth the variance components model (no lower bound of zero on the estimate) the ADE and ACE models fit equally well, though typically only one will give plausible non-negative estimates, unless rMZ=2rDZ so both C and D are estimated at zero.
*Thread Reply:* Great, thanks Michael, this is greatly appreciated! I'll have a look at Ben's talk again as the dominance effects still puzzles me a bit 😬
3/ 4 When testing the significance of the estimates in univariate nested models: how can one interpret if both A and E are not significant (AE vs ACE, CE vs ACE) but simultaneously A & C are significant (E vs ACE)?
4 / 4 what can one do if the assumptions in the twin data are violated, ie mean and variances are not equal either between twin order or between zygosity groups?
Hi! I'm not sure if anyone is still reading slack messages, but I seem to be having an issue when trying to run @Hermine Maes oneSATc.R script in OpenMx. The instructions in the video are very helpful and clear; however, when I attempt to run the saturated model " fitSAT <- mxRun( modelSAT, intervals=F ) ", I am receiving an error "Error: 'is_present' is not an exported object from 'namespace:lifecycle' ". I haven't encountered an error until this point in the script. I am still somewhat new to R, so I could be missing something very obvious, but any assistance would be helpful! Thank you!
*Thread Reply:* no idea what this means, but I’ll let Hermine know about your question
*Thread Reply:* I’m not sure why you’re getting this error as it runs fine here. What version of R and OpenMx are you running?
*Thread Reply:* It’s possible that the lifecycle package is out of date. Please try the following:
*Thread Reply:* install.packages(“lifecycle”)